Overview
Dr. Young laboratory’s primary interest is the study of lymphoma, myeloma and other primary lymphoproliferative disorders. We focus on identification of specific biomarkers and molecular mechanisms for accurate prognostic prediction, sub-classification, and therapeutic targets in lymphoid neoplasms and diseases. Our contributions on the p53 pathway and MYC/BCL2 double-hit and double-expressors in diffuse large B-cell lymphoma are well-recognized by the Hematology and Oncology, and Pathology Societies. The long-term goal of Dr. Young’s team is to focus on the translational research with enthusiasm to develop biomarker-guided novel therapies to improve the prognosis, prediction, treatment response and possibly to overcome the drug resistance.
Research Interests
The Young laboratory’s major focus is the study of B-cell lymphoma and myeloma. We are most interested in genetic, epigenetic, and immunological mechanisms underlying the heterogeneous clinical outcome of diffuse large B-cell lymphoma (DLBCL) and high-grade B-cell lymphoma (HGBCL). We used a variety of research approaches to gain insights into the molecular abnormalities at the DNA, RNA, and protein levels. We have contributed to the identification of several biomarkers recognized by the WHO classification of lymphoid neoplasms (e.g., MYC/BCL2 genetic double-hit and phenotypic double-expressors, DLBCL subtyping IHC algorithm, CD5+, CD30+, HIV+ and EBV+). We make efforts in risk stratification and compare different therapies in rare subtypes of lymphomas including Castleman lymphadenopathy and Waldenström macroglobulinemia, and have provided important clinical data to the community of clinicians and scientists for making clinical decisions and future therapeutic studies.
1. DLBCL or HGBCL is the most common type of aggressive B-cell lymphoma. Nearly 70% DLBCL patients are cured with the standard therapyrituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP), whereas patients who are refractory or relapsed disease have low survival rates after treated with salvage chemotherapy followed by high-dose therapy and autologous stem cell transplant. The substantial variations in clinical outcome of DLBCL are attributable to the high heterogeneity of DLBCL pathogenic mechanisms. Figure below shows the identified genetic alterations in p53 and MYC pathways in DLBCL as we summarized in Nat Rev Clin Oncol 2019;16(10) by Miao and Young, et al. Current molecular sub-classification of DLBCL is based on the cell-of-origin expression signatures of DLBCL gene expression profiles at the RNA level. In recent years, novel genetic models based on pathogenic alterations are being developed. Recently we have published our novel DLBCL classifiers using combined genetic and gene-expression signatures constructed by Artificial Intelligence (Xu-Monette et al, Blood Adv. 2020; Oncoimmunotherapy 2021; CCR 2022).
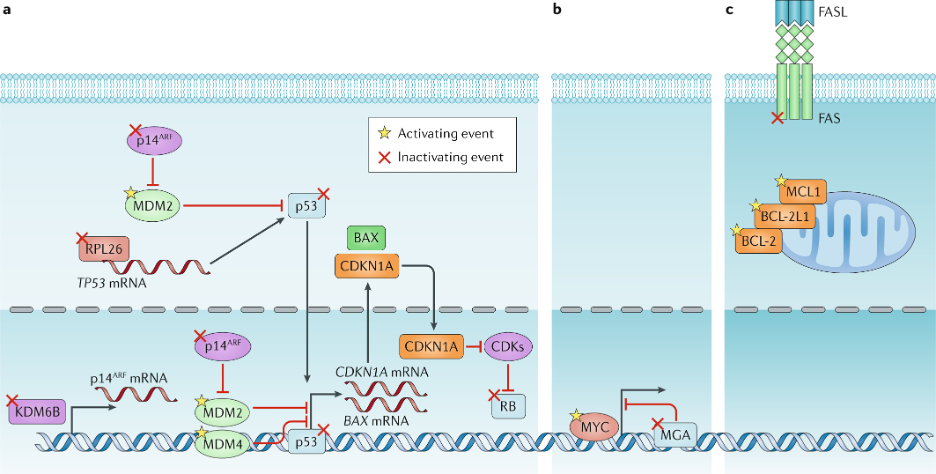
2. In addition to tumor-intrinsic mechanisms underlying the heterogeneous clinical outcome of DLBCL, we have also investigated the dysregulation of tumor immune microenvironment using DLBCL diagnostic samples. Our publication in Cancer Immunol Res. 2019 by Xu-Monette et al is the first large-scale immune profiling study in DLBCL using the state-of-the-art fluorescent multiplex immunohistochemistry and automated quantitation technology, which can quantitate cell-type specific expression with high-sensitivity. We are continuing to identify immunosuppressive mechanisms in DLBCL with clinical significance.
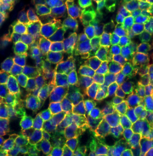
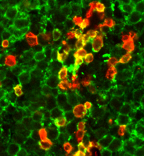
For two preceding figures: (Xu-Monette ZY, Young KH, et al. Immune profiling and quantitative analysis decipher the clinical role of immune-checkpoint expression in the tumor immune microenvironment of DLBCL. Cancer Immunol Res. 2019;7(4):644-657.)
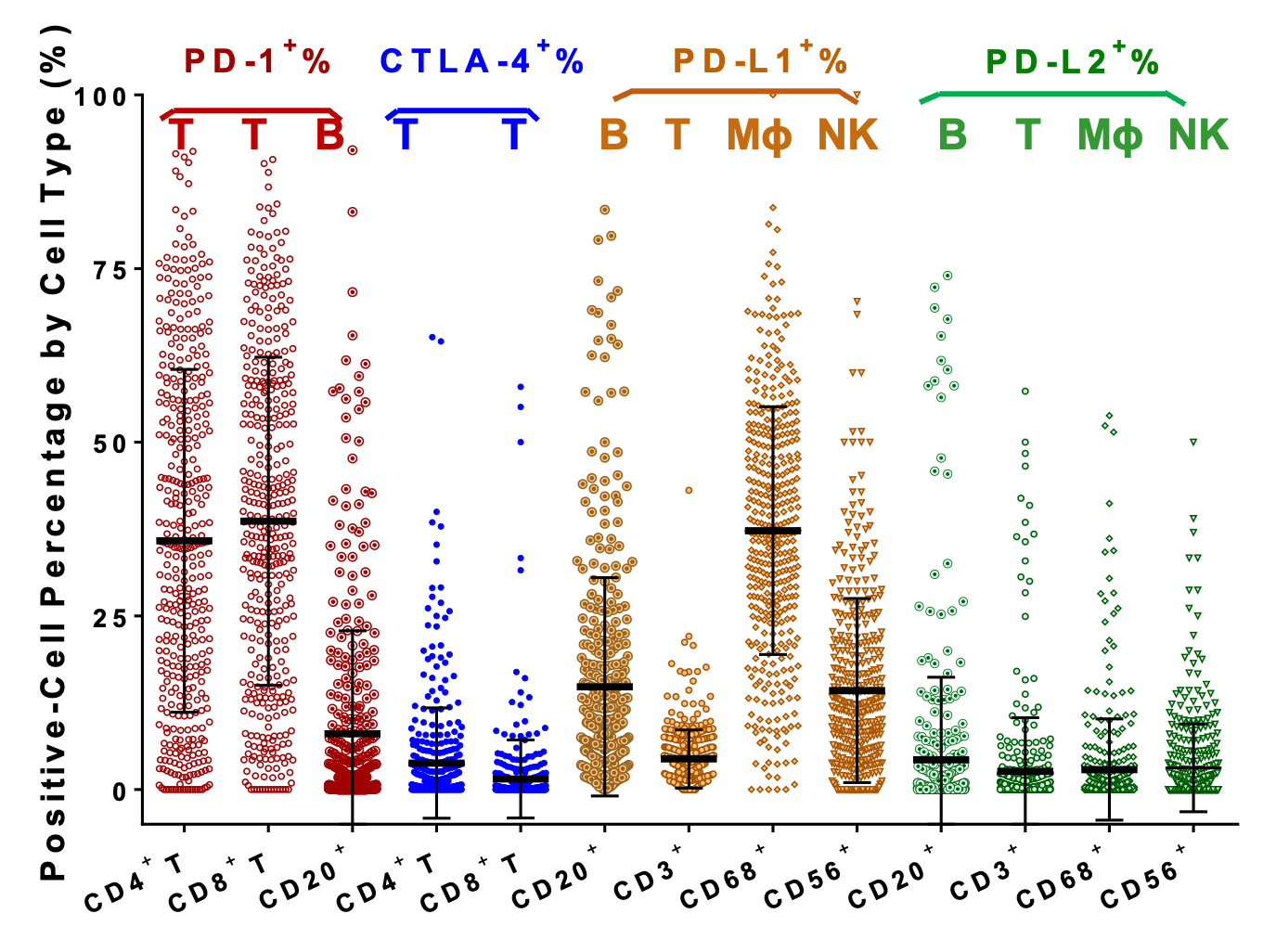
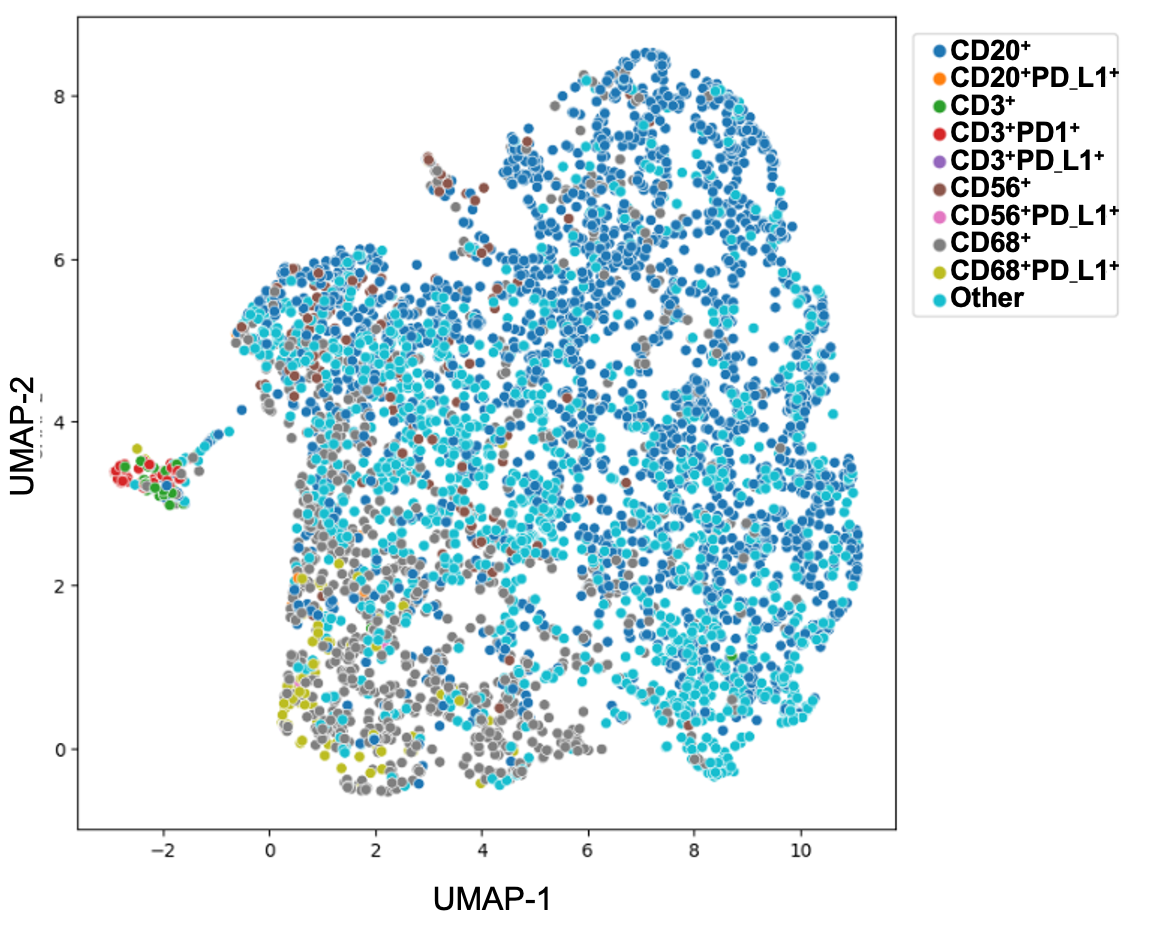
For two preceding figures: (Xu-Monette ZY, Young KH, et al. Immune profiling and quantitative analysis decipher the clinical role of immune-checkpoint expression in the tumor immune microenvironment of DLBCL. Cancer Immunol Res. 2019;7(4):644-657.)